We highlight the research initiatives that define our Lab’s focus and expertise. Each project reflects our commitment to translational microbiome research and host–microbe interactions, combining experimental and computational approaches to advance scientific understanding and translational applications. Explore the projects below to learn about our ongoing studies, methodologies, and the impact of our work. We hope this provides insight into how our team collaborates, innovates, and contributes to the broader scientific community.
MICROBENCH
Microbiota reproducibility benchmark project
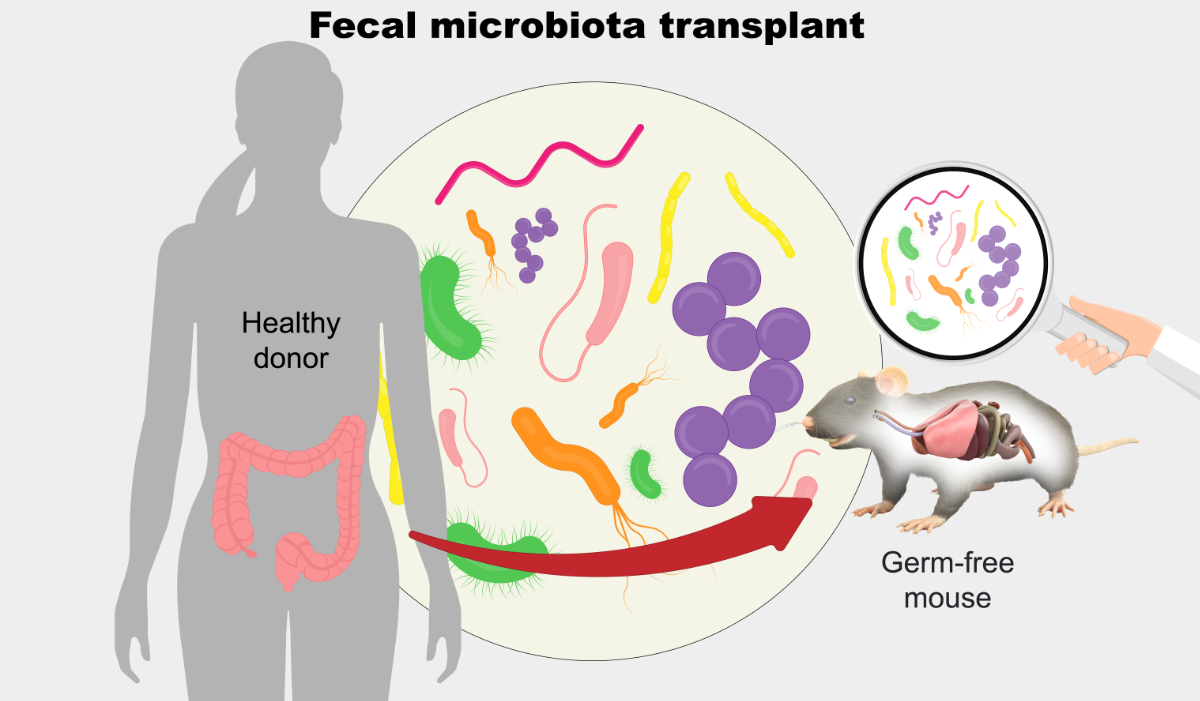
Background
Human microbiota-associated (HMA) mice enable controlled testing of how human gut microbes influence host biology. However, engraftment outcomes are highly variable: many human taxa fail to persist in mice, and microbial communities often diverge substantially from their donors. Methodological differences—such as donor handling, colonization protocols, and timing of sampling—further reduce reproducibility. Despite widespread reports of successful phenotype transfer, these inconsistencies raise concerns regarding experimental and statistical rigor.
Objective
This project systematically evaluates the technical and biological factors that determine successful, reproducible engraftment. By integrating longitudinal microbiome and host measurements, we aim to establish evidence-based best practices for generating and validating HMA models.
Aims
- Quantify engraftment efficiency across experimental conditions, including donor variability, recipient factors, and time after colonization, using Oxford Nanopore sequencing.
- Identify procedural determinants—such as inoculum preparation, storage conditions, and sampling schedules—that improve microbial and phenotypic reproducibility across replicates.
- Develop and share a standardized, empirically validated protocol for generating and quality-controlling HMA mice to improve transparency, reproducibility, and interpretation across microbiome studies.
Through this work, we aim to establish a quantitative framework linking experimental design to engraftment outcomes. The resulting guidelines will support more rigorous use of HMA models, improve cross-study comparability, and facilitate more reliable discovery of microbiome-driven mechanisms.